Professor Sir Alan Fersht
Scientific Advisory Board Chair

Pioneering protein folding professor
The University of Cambridge
Ranked in the top 100 UK Scientists. Sir Alan Fersht’s current research at The University of Cambridge uses an amalgam of protein engineering, structural biology, biophysics and chemistry to study the structure, activity, stability and folding of proteins, and the role of protein misfolding and instability in cancer and disease. His group at the MRC Laboratory of Molecular Biology in Cambridge focuses on how mutation affects proteins in the cell cycle, particularly the tumour suppressor p53, in order to design novel anti-cancer drugs that function by restoring the activity of mutated proteins. Since October 2012-2018, he has been Master of Gonville and Caius College, Cambridge. Knighted in 2003 for his work on protein science, and has honorary degrees from Uppsala, Brussels, Weizmann Institute, Imperial College and The Hebrew University. He has honorary professorships in China in Shanghai, Chengdu and Qingdao. He has world-wide experience in the needs of science and biotechnology.
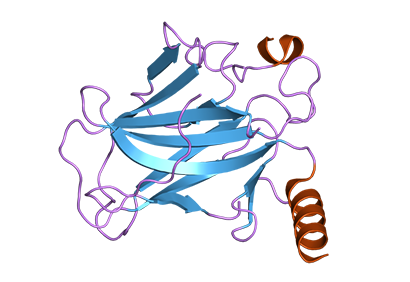
Multisite aggregation of p53 and implications for drug rescue
Protein aggregation is involved in many diseases. Often, a unique aggregation-prone sequence polymerizes to form regular fibrils. Many oncogenic mutants of the tumor suppressor p53 rapidly aggregate but form amorphous fibrils. A peptide surrounding Ile254 is proposed to be the aggregation-driving sequence in cells. We identified several different aggregating sites from limited proteolysis of harvested aggregates and effects of mutations on kinetics and products of aggregation. We present a model whereby the amorphous nature of the aggregates results from multisite branching of polymerization after slow unfolding of the protein, which may be a common feature of aggregation of large proteins.
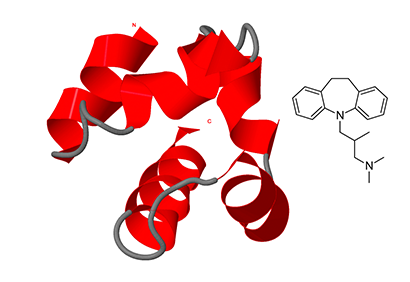
Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm
We have previously shown that the thermolabile, cavity-creating p53 cancer mutant Y220C can be reactivated by small-molecule stabilizers. In our ongoing efforts to unearth druggable variants of the p53 mutome, we have now analyzed the effects of other cancer-associated mutations at codon 220 on the structure, stability, and dynamics of the p53 DNA-binding domain (DBD)….Some of the compounds also bound to and stabilized the Y220N mutant, but not the Y220H mutant. Our data validate the Y220S and Y220N mutants as druggable targets and provide a framework for the design of Y220S or Y220N-specific compounds as well as compounds with dual Y220C/Y220S specificity for use in personalized cancer therapy.

The Selected Papers of Sir Alan Fersht: Development of Protein Engineering
The publication of the ‘The Selected Papers of Sir Alan Fersht’ is an invitation to look back over a fabulously productive scientific career. The book reproduces 48 papers beginning with Fersht’s first publication as a PhD student on the reactions of aspirins and follows the progression of his work into enzyme mechanism, protein engineering and protein folding, and on to protein stability and cancer. As Alan Fersht has authored over 500 publications, the papers presented are indeed select. They are grouped logically to reflect the themes of the research and the phases of Alan’s career. Each group of papers is introduced with entertaining and informative commentaries rich in opinion, historical context, personal reminiscences and anecdotes and contains a collection of photographs of colleagues and coworkers. Although Alan is most closely associated with Cambridge, he spent 10 productive years at Imperial College London and the college is to be congratulated on publishing this volume of Selected Papers.